|

1) Building a Kinase Active-Site Peptide Library
단백질 데이터베이스로 부터 꾸준하게 단백질 활성효소의 Sequence와 활성자리가 알려진 반면에 검출 및 확인, 정량분석을 위한 방법을 만드는 것은 여전히 도전적인 일이다. 이러한 펩타이드들의 대다수가 그들 각자의 활성효소들에 대하여 유일무이하기 때문에 본 실험 방법은 활성효소의 활성자리 펩타이드들을 정성 분석하고 정량 분석하는 것에 중점을 두고 있다. 더불어, 이러한 펩타이드들은 다양한 활성효소 영역을 가진 활성효소들에 대하여 활성자리 억제에 관하여 직접적인 통찰을 규정한다.
활성효소의 활성자리 펩타이드들에 대한 목록을 만들기 위해서는 활성자리 펩타이드를 농후화시키기 위한 미처리 세포 용해물 샘플들을 ActivX Desthiobiotin-ATP 또는 -ADP Probe로 표시한다. 초기에, Proteome Discoverer Software을 사용하여 데이터베이스 검색을 위한 스펙트라 라이브러리 생성을 위하여 일반 모드로 TOP 10 데이터 획득 방법(Table 1)을 사용하였다. 이러한 검색의 결과들은 펩타이드 Sequence와 Desthiobiotin 변형 자리 그리고 단백질 활성효소의 구성원을 결정하는데 사용된다. 더불어, 이러한 실험방법은 펩타이드의 Retention Times 과 Precursor 그리고 Product Ion의 Charge States, HCD Product Ion의 분포 등을 포함하는 그 다음의 Targeted Acquisition 방법들에 중요한 데이터를 제공했다.
펩타이드 Sequence는 단백질의 기능을 확인하기 위하여 Thermo Scientific Protein Center™ Software을 사용하였다. Figure 2는 Desthiobiotin-ATP 또는 –ADP Probe로 표시한 펩타이드로부터 분석된 단백질들을 보여준다. 두 Probe를 사용하여 식별된 총 단백질의 13%을 대표하는 단백질 활성효소와 알고있는 표시된 ATP 단백질들(각각 77%, 83%)에 대하여 두 Probe들을 모두 높은 특이성을 보였다. 비록 두 Probe가 비슷한 수의 활성효소들을 보통 정도의 중복된 상태로 농후화시켰을지라도, 전에 보고된 바와 같이 특정한 활성효소에 대한 각각의 Probe들의 선호하는 결합이 있다. Figure 3A는 Tao1/3의 Targeted 활성자리 펩타이드 DIKAGNILLTEPGQVK의 Retention Time에서의 HR/AM MS 스펙트럼을 보여주고 있다.
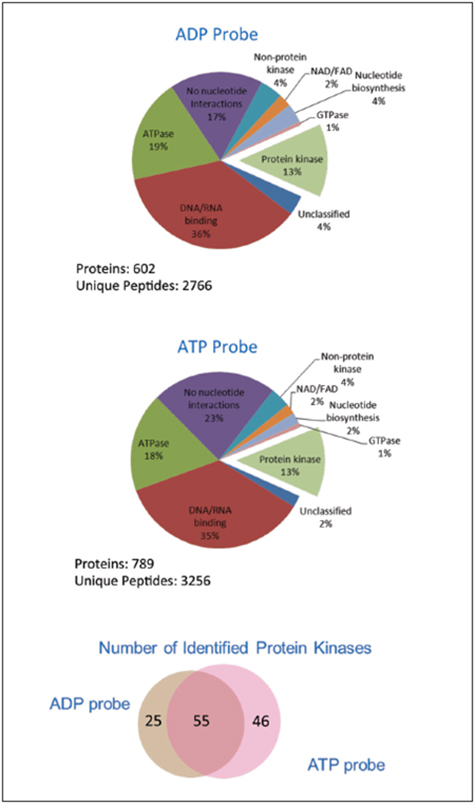
Figure 2 . Proteins identified after desthiobiotin-ATP and –ADP probe labeling and enrichment categorized by protein Function using Protein Center software. The Venn diagram shows the distribution of the resulting protein kinases identified Using each probe.
복잡한 스펙트럼들은 전형적으로 활성자리 펩타이드들을 농후화시킨 후에 나타나고, Multiple Charge 띤 다양한 펩타이드들을 함유하고 있다는 것이다. 순차적인 HR/AM MS와 MS/MS 획득을 위한 Q Exactive의 속도는 Orbitrap Mass Analyzer에서 C-trap의 이온 농도 증가와 조화를 이루어 +3 Precursor가 14번째로 많이 있는 펩타이드 임에도 불구하고 +3 Precursor를 일반 모드에서 선택하고 Sequencing할 수 있었다. Figure 3B는 +3 Charge State를 갖는 DIKAGNILLTEPGQVK Precursor에 대하여 실험적으로 측정된 Isotopic의 분포와 이론적인 Isotopic의 분포의 상응을 보여준다. 각각의 Isotopic은 이론적인 Isotopic의 Intensity 분포와 비교하였을 때 5 ppm미만 그리고 15% 미만의 Mass Error를 갖는다. Figure 3C는 25 b- 그리고 y-type Fragment 이온들과 대조한 HCD MS/MS 스펙트라 데이터베이스 검색결과를 보여준다.
Fragment 이온들에 대한 Mass Error의 평균값은 측정된 Product Ion Intensities의 전체 Order of Magnitude Range에 대해 2 ppm미만이었다. Mass 스펙트럼 전체에 걸쳐 변함없는 Mass Error를 유지하는 능력은 펩타이드 분석의 신뢰성을 매우 증가시킬 수 있기 때문에 Orbitrap을 이용한 검출에 있어 탁월한 장점이라고 할 수 있다.
궁극적으로, Proteome Discoverer Software로 분석된 126 활성효소 활성자리 펩타이드들은 상대적인 양을 평가하고 예정된 목적물질 획득을 위한 Inclusion List를 만들기 위해 Pinpoint Software로 보내어진다(Figure 3D).
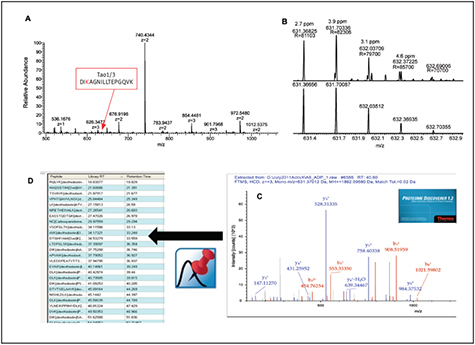
Figure 3. Processing strategy for building kinase active-site peptides spectral libraries and target lists. Example active-site peptide DIKAGNILLTEPGQVK from Tao1/3 kinase (A, B) was identified using Proteome Discoverer software from the HCD product ion spectrum (C) with Pinpoint software used to automate target-list method building (D).
|