
Solvent Sample Analysis: The results showed a distinct difference between the spiked
and controlled blank samples (Figure 4 ). The ChemSpider search returned with the
correct identification of three compounds: Strychnine, Colchicine and Aconitine with the
mass tolerance of 2 ppm with external calibration (Figures 5 & 6). Notice that two of the
spiked toxins, Colchicine and Aconitine (Figure 6), overlapped with each other in
UHPLC chromatograms with a short gradient but were accurately identified by SIEVE
software via a differential analysis between samples versus the control blank when
searched against KEGG and Sigma-Aldrich online databases in ChemSpider.
FIGURE 4. Chromatographic Alignment, Frame and PCA Analysis of Solvent Sample1 Spiked with 3 Toxins
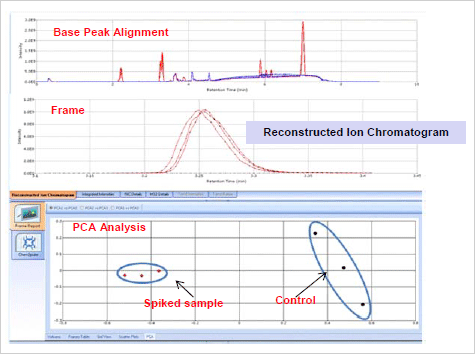
FIGURE 5. Identified Toxin1-Strychnine(m/z 335.1751) in Solvent Sample
FIGURE 6. Identified Toxin2-Colchicine(m/z 400.1749) and Toxin3-Aconitine (m/z 646.3215) in Solvent Sample
Apple Juice Sample Analysis: Two toxins were spiked in the apple juice sample.
Three replicates were analyzed against a clean apple juice matrix. Results are
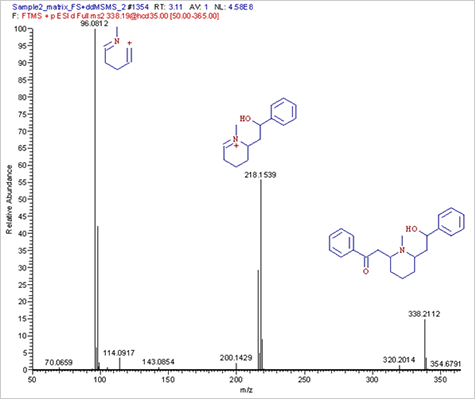
shown in Figures 7,8 & 10. Lobeline was fairly easily identified as the first spiked
toxin (Figure 8). The identity of Lobeline was confirmed by MSMS interpretation as
shown in Figure 9.
FIGURE 7. Apple Juice Sample-PCA Analysis
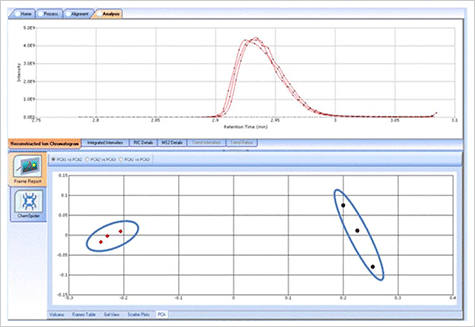
FIGURE 8. Identified Toxin1-Lobeline(m/z 338.2113) in Apple Juice Sample
FIGURE 9. Annotated MS/MS spectrum of Lobeline(m/z 338.2113)
In apple juice sample, Solanine m/z 868.5031 and its in-source fragmentation ions,
Solanadine m/z 398.3410, 560.3936, 561.3970 (n+1 isotope of 560.3936) co-eluted at
3.26 min and were identified as the 2nd toxic compound (Figures10 & 11).
FIGURE 10. Identification of Toxin2-Solanine(m/z 868.5031) and its In-Source
Fragment Solanadine(m/z 398.3410) in Apple Juice Sample
FIGURE 11. MS/MS Spectrum of m/z 868.5031

| • |
Acetonitrile solvent sample spiked with 3 toxins, apple juice sample with 2 toxins and
matrix blanks were analyzed in triplicate by the Q Exactive MS with Full Scan at
70,000 and Top1 MS/MS at 17500 resolution. |
|
| • |
Mass accuracy of all identified toxins is within 2 ppm with external mass calibration. |
|
| • |
Spiked toxins were screened through SIEVE1.3 software. Four out of five unknowns
were easily and successfully targeted. The other toxin was found to co-eluted with its
in-source fragmentation products. |
|
| • |
With the function of precursor ion selection for MS/MS and the HR/AM data acquired
from the Q Exactive MS, unknown compounds can be screened out at high
confidence with the structure elucidation within one injection. |
ChemSpider is a trademark of ChemZoo Inc. and RSC Worldwide Limited. All other trademarks are the property of
Thermo Fisher Scientific Inc. and its subsidiaries. This information is not intended to encourage use of these products in
any manners that might infringe the intellectual property rights of others.
|